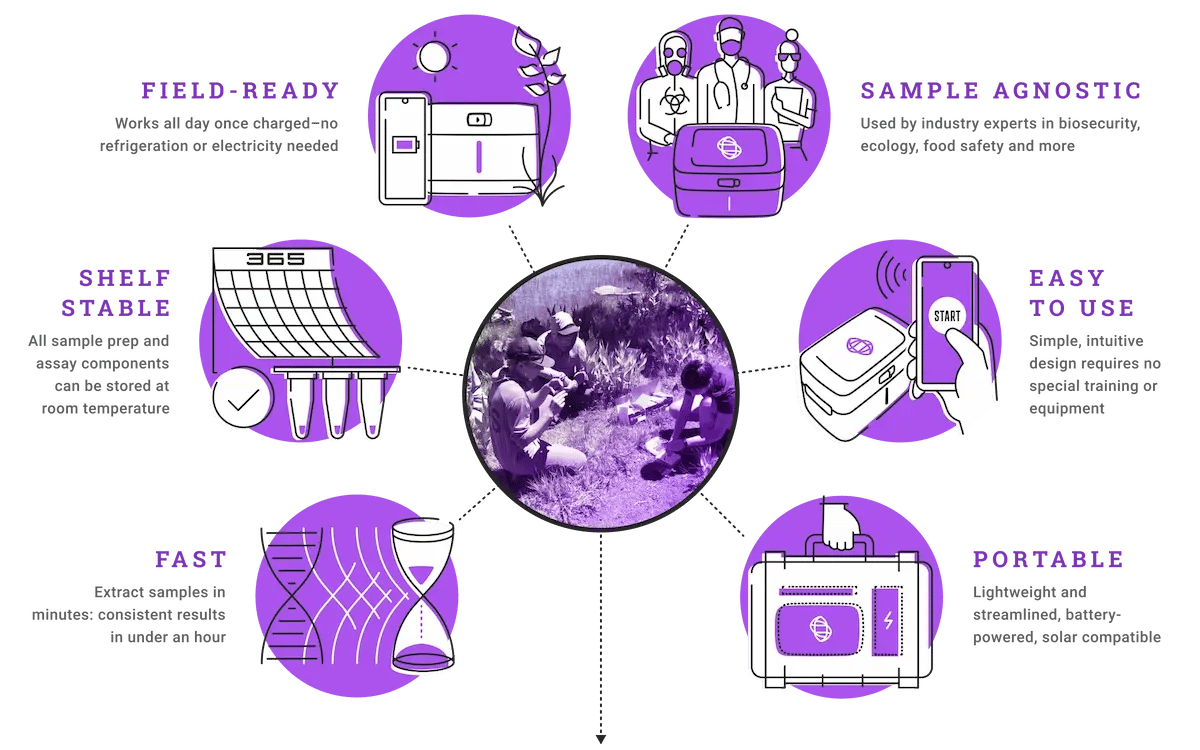
From protecting global health to preparing for the next pandemic, our groundbreaking molecular testing platforms and products are essential equipment for field researchers in any ecosystem across the planet.

Equip yourself with powerful tools that can handle any research challenge. Our platforms allow you to analyze various sample types for multiple targets with ease, no matter where you are. Experience unmatched speed, sensitivity, and reliability that redefines what’s possible in the field.
Bring your research into the field–our versatile, fully mobile lab goes to any sample site with ease. Researchers use Biomeme's deployable system for rapid, gold standard detection and quantification across various matrices and environmental settings.
Rapid detection and response for when every minute matters
Get reliable, actionable results in real-time
Versatile, sensitive tests save time, money, and labor
Field-friendly for the unfriendliest conditions, our fully mobile, easy to use products and platforms provide rapid results that you can trust every time.







After 15 years working in molecular diagnostics, I know how far the science has come and who is doing what in this...
401 North Broad St Suite 222 Philadelphia, PA 19108